In the paper “Krishna, G., Khan, K., & Roy, K. (2020). Application of QSARs in identification of mutagenicity mechanisms of nitro and amino aromatic compounds against Salmonella typhimurium species. Toxicology in Vitro, 65(November 2019), 104768. https://doi.org/10.1016/j.tiv.2020.104768“, the authors presented QSAR models to predict mutagenicity mechanisms of nitro and amino aromatic compounds against Salmonella typhimurium species.
The information described in the paper was used to build two alvaRunner projects that we present here. The models were created using the paper’s datasets.
alvaRunner projects
Two alvaRunner projects are available. The first one includes the ordinary least squares (OLS) models based on TA98-S9 dataset. The TA98-S9 dataset has been curated using alvaMolecule, ten molecules included in the original dataset have been identified as wrong and fixed in accordance with the authors of the paper.
The second alvaRunner project includes the models based on TA98+S9 dataset.
TA98-S9 project
The TA98-S9 project contains four regression models:
- TA98-S9 Model-1: an ordinary least squares model based on 8 molecular descriptors (MD) built using the 219 molecules of the paper training set
- TA98-S9 Model-2: an ordinary least squares model based on 8 molecular descriptors (MD) built using the 219 molecules of the paper training set
- TA98-S9 Model-1 All: an ordinary least squares model based on TA98-S9 Model-1 descriptors built using all the 291 molecules
- TA98-S9 Model-2 All: an ordinary least squares model based on TA98-S9 Model-2 descriptors built using all the 291 molecules
Models TA98-S9 Model-1 and TA98-S9 Model-1 All include the following eight molecular descriptors:
- nCIR: number of circuits
- NRS: number of ring systems
- X5Av: average valence connectivity index of order 5
- RDCHI: reciprocal distance sum Randic-like index
- Eta_D_beta: eta measure of electronic features (corresponds to the ETA_dBeta descriptors cited in the paper)
- B08[Cl-Cl]: Presence/absence of Cl – Cl at topological distance 8
- F07[C-C]: Frequency of C – C at topological distance 7
- F09[N-N]: Frequency of N – N at topological distance 9
Models TA98-S9 Model-2 and TA98-S9 Model-2 All include the following eight molecular descriptors:
- RCI: ring complexity index
- NRS: number of ring systems
- X5A: average connectivity index of order 5
- RDCHI: reciprocal distance sum Randic-like index
- Eta_betaP: eta pi and lone pair VEM count (corresponds to the ETA_Beta_ns descriptors cited in the paper)
- Eta_D_beta: eta measure of electronic features (corresponds to the ETA_dBeta descriptors cited in the paper)
- B09[N-N]: Presence/absence of N – N at topological distance 9
- F07[C-C]: Frequency of C – C at topological distance 7
Since RDCHI descriptor values increases with the size of the molecules whereas it decreases with branching, the authors suggest that higher values of this descriptor produces more toxic, potentially more mutagenic nitro-aromatic compounds. ETA_D_beta is related to the relative unsaturation content of the molecular structure, it contributes positively towards the mutagenicity, i.e., the mutagenicity of the nitro-aromatic compounds increases with an increase of unsaturation in the form of double bonds. ETA_betaP is a measure of electron richness in a molecule, this index has a negative effect towards mutagenicity, maybe due to an increase in polar bulk in the molecule. nCIR, RCI and NRS descriptors suggest that mutagenicity can be influenced by the presence of various ring systems. X5A and X5AV positively influence the mutagenicity, high values of these descriptors correspond to an increase in size and non-polar surface area of the molecule, suggesting that mutagenicity may increase with an increase in the surface area and size of the molecules.
The scores of the models of the alvaRunner project are presented in the following table:
| Model name | Training | Test | ||||
|---|---|---|---|---|---|---|
| R2 | Q2CV | RMSE | RMSECV | R2 | RMSE | |
| TA98-S9 Model-1 | 0.729 | 0.703 | 1.009 | 1.055 | 0.739 | 0.938 |
| TA98-S9 Model-2 | 0.714 | 0.682 | 1.035 | 1.092 | 0.728 | 0.957 |
| TA98-S9 Model-1 All | 0.734 | 0.722 | 0.989 | 1.011 | – | – |
| TA98-S9 Model-2 All | 0.721 | 0.702 | 1.013 | 1.046 | – | – |
The following charts show the predicted (Y) and real (X) values of the models:
| TA98-S9 Model-1 | TA98-S9 Model-2 |
|---|---|
 |
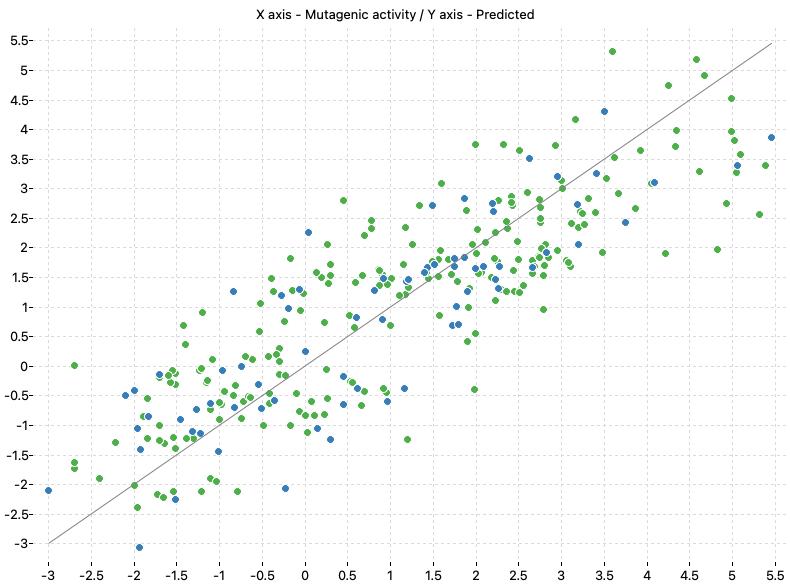 |
TA98+S9 project
The TA98+S9 project contains four regression models:
- TA98+S9 Model-1: an ordinary least squares model based on 8 molecular descriptors (MD) built using the 232 molecules of the paper training set
- TA98+S9 Model-2: an ordinary least squares model based on 8 molecular descriptors (MD) built using the 232 molecules of the paper training set
- TA98+S9 Model-1 All: an ordinary least squares model based on TA98+S9 Model-1 descriptors built using all the 309 molecules
- TA98+S9 Model-2 All: an ordinary least squares model based on TA98+S9 Model-2 descriptors built using all the 309 molecules
Models TA98+S9 Model-1 and TA98+S9 Model-1 All include the following eight molecular descriptors:
- D/Dtr09: distance/detour ring index of order 9
- Eta_epsi_3: eta electronegativity measure 3 (corresponds to the ETA_Epsilon_3 descriptors cited in the paper)
- nPyridines: number of Pyridines
- C-034: R–CR..X
- SsssCH: Sum of sssCH E-states
- SaaaC: Sum of aaaC E-states
- B06[C-C]: Presence/absence of C – C at topological distance 6
- F02[N-N]: Frequency of N – N at topological distance 2
Models TA98+S9 Model-2and TA98+S9 Model-2 All include the following eight molecular descriptors:
- Eta_epsi_3: eta electronegativity measure 3 (corresponds to the ETA_Epsilon_3 descriptors cited in the paper)
- nImidazoles: number of Imidazoles
- nPyridines: number of Pyridines
- C-027: R–CH–X
- SsssCH: Sum of sssCH E-states
- SaaaC: Sum of aaaC E-states
- SaaNH: Sum of aaNH E-states
- B02[N-N]: Presence/absence of N – N at topological distance 2
The authors highlighted the importance of Eta_epsi_3, suggesting that with an increase in the electronegative element content (mainly nitrogen), the tendency of molecules to behave as potent mutagenic entity is enhanced in aromatic and hetero-aromatic compounds. nImidazoles, F02[N-N], B02[N-N] and SaaNH are related to the presence of high electronegative element content in the aromatic or hetero-aromatic amine datasets. SaaaC, B06[C-C] and D/Dtr09 can be related to the lipophilic bulk of the organic
chemicals.
The scores of the models of the alvaRunner project are presented in the following table:
| Model name | Training | Test | ||||
|---|---|---|---|---|---|---|
| R2 | Q2CV | RMSE | RMSECV | R2 | RMSE | |
| TA98+S9 Model-1 | 0.701 | 0.667 | 0.909 | 0.959 | 0.696 | 0.896 |
| TA98+S9 Model-2 | 0.692 | 0.634 | 0.922 | 1.006 | 0.662 | 0.946 |
| TA98+S9 Model-1 All | 0.701 | 0.689 | 0.905 | 0.922 | – | – |
| TA98+S9 Model-2 All | 0.687 | 0.667 | 0.926 | 0.955 | – | – |
The following charts show the predicted (Y) and real (X) values of the models:
| TA98+S9 Model-1 | TA98+S9 Model-2 |
|---|---|
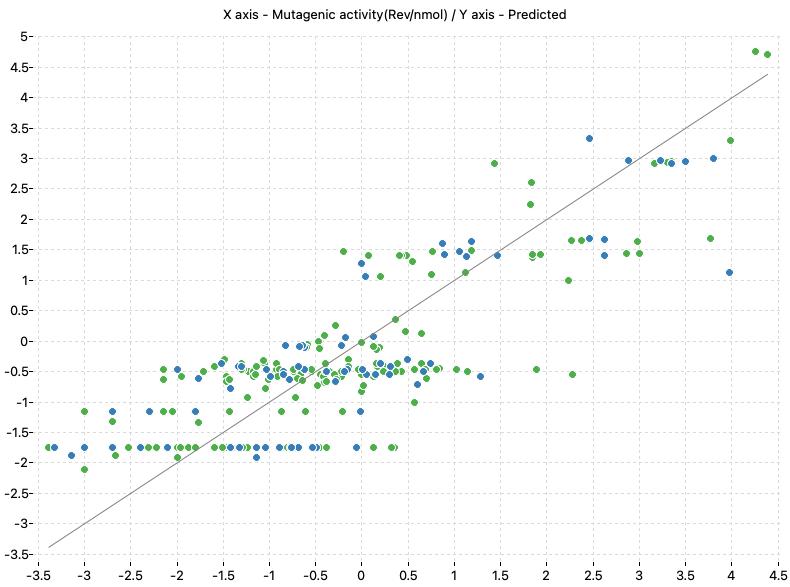 |
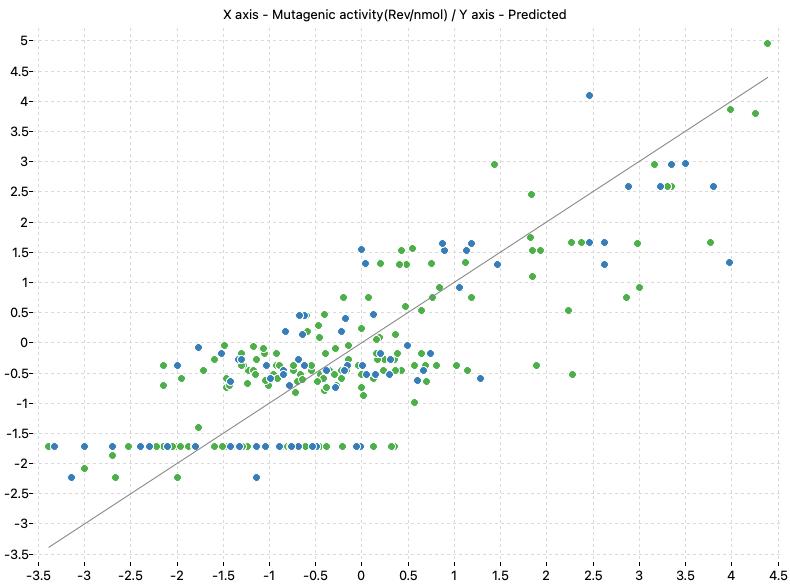 |
